324
Views & Citations10
Likes & Shares
Human infections occur through the consumption of contaminated food such as undercooked meat, drinking contaminated water, or direct person-to-person contact. In developing countries, like Nigeria, the major causes of infantile diarrhea are ETEC, EPEC and EAEC, while EHEC and EAEC are mainly associated with food poisoning, i.e. cooked and uncooked food. The presence of intestinal and extraintestinal pathogenic E. coli in food suggests a public health impact and inconsistency health hazard, and agencies such as the NAFDAC in Nigeria report food borne outbreaks associated with verotoxigenic (VTEC) and another pathogenic E. coli. There should be a concerted effort in reducing the microbial menace of E. coli strains with the enhanced general awareness of VTEC following reports of large outbreaks in the African country and worldwide [4].
The virulent gene understudy in this research work are 1, (eaeA gene), 2 (easTgene) and 3 (bfpgene) and the ability of virulence and resistance genes were believed to increase the degree pathogenicity of a microorganism and the prevalence of infection with the great possibility of therapy failure. Escherichia coli is an opportunistic pathogen, commensal bacteria that can be found as normal flora in humans and animals and contains the inherent ability of this virulent gene, eaeA gene, easTgene, and bfpgene. A subset of these genes will be key players in the ability of the bacterium to cause disease. The products of such genes that facilitate the successful colonization and survival of the bacterium in or cause damage to the host are considered as virulence or pathogenicity determinants. This is one of the pre-determinant factors to E. coli ability to cause infection [5].
The east gene is expressed in the neurogenic region at the time of neuroblast segregation and in cells in the peripheral and central nervous system, bfpgene is the Blue fluorescent protein derived from the mutated purple chromoprotein isolated from the sea anemone Stichodactyla haddoni and easTgene while eaeA, a gene necessary for the characteristic intimate attachment of EPEC to epithelial cells. is the focal point of this research work, all together constitute the pathogenic factor and function for E. coli pathogenic tendencies.
- coli bacteria in their intestines can get on the meat. Slaughtered Beef combines meat from many different animals, increasing the risk of contamination with E. coli. While most people experience a few days of upset stomach and then recover fully, E. coli infections can sometimes be life-threatening [6].
This research work will seek to educate the Nigerian populate and worldwide as a whole and deplete that the most way to get an E. coli infection is by eating contaminated food, such as: Slaughtered Beef from contaminated Slaughtered houses and other beef handler. When cattle are slaughtered and processed.
MATERIALS AND METHODS
Bacteria isolates
This research employed 15 E. coli isolates coded as samples AA1, AA3-2, AA-3, A2, A1-2, A2-2, A4-2, A1-3, A4-3, A1-2, A2-2, A2-3, AA-2, and A1-3. The bacterial isolates were obtained from 48 slaughter beef samples from slaughter house in several Local markets in Akungba Akoko, Ondo State, Nigeria. There were 12 meat samples from the Akoko North market (ASAN) and 6 isolates, 26 meat samples from Akoko south market (AAS) and 2 isolates, 6 meat samples from the Akoko East market (AAE) and 5 isolates, and 4 meat samples from the Akoko West market (AAW) and 2 isolates were found. These isolates were screamed for the presence of target virulent gene and identified as pathogenic E. coli [7,8].
Cultivation of bacteria isolates
These fifteen isolates were taken from isolate stock kept in a freezer at -20°C. These were then grown in Brain Heart Infusion Broth (BHIB) media and incubated for 24 h at 37°C. Bacterial growth was evident with increasing turbidity of BHIB media [8,9].
DNA extraction of bacteria isolates
DNA was extracted using the protocol stated by Osuntokun [10]. DNA was extracted using the protocol stated by Pitout [1]. Briefly, Single colonies grown on medium were transferred to 1.5 ml of liquid medium and cultures were grown on a shaker for 48 h at 28ºC. After this period, cultures were centrifuged at 4600g for 5 min. The resulting pellets were resuspended in 520 μl of TE buffer (10 mM Tris-HCl, 1mM EDTA, pH 8.0). Fifteen microliters of 20% SDS and 3 μl of Proteinase K (20 mg/ml) were then added. The mixture was incubated for 1 hour at 37ºC, then 100 μl of 5 M NaCl and 80 μL of a 10% CTAB solution in 0.7 M NaCl were added and votexed. The suspension was incubated for 10 min at 65ºC and kept on ice for 15 min. An equal volume of chloroform: isoamyl alcohol (24:1) was added, followed by incubation on ice for 5 min and centrifugation at 7200g for 20 min. The aqueous phase was then transferred to a new tube and isopropanol (1: 0.6) was added and DNA precipitated at -20ºC for 16 h. DNA was collected by centrifugation at 13000g for 10 min, washed with 500 μl of 70% ethanol, air-dried at room temperature for approximately three hours and finally dissolved in 50 μl of TE buffer [9,11].
Molecular identification of bacteria isolates
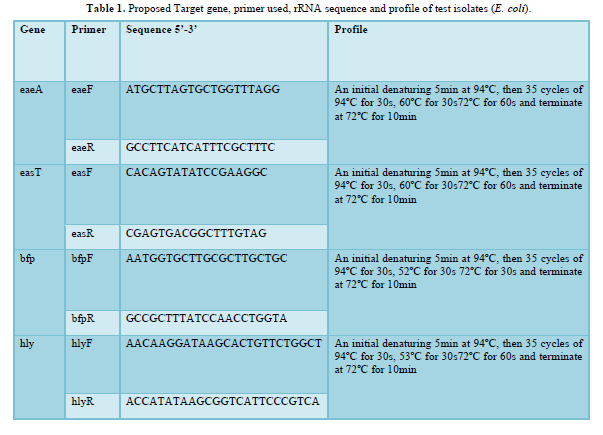
Primer sequences were as earlier documented. Reaction cocktail used for all PCR per primer set included (Reagent Volume µl) - 5X PCR SYBR green buffer (2.5), MgCl2 (0.75), 10pM DNTP (0.25), 10pM of each forward and backwards primer (0.25), 8000U of taq DNA polymerase (0.06) and made up to 10.5 with sterile distilled water to which 2µl template was added. Buffer control was also added to eliminate any probability of false amplification Table below shows the primer sequence and PCR profile used in amplifying each fragment. PCR was carried out in a GeneAmp 9700 PCR System Thermalcycler (Applied Biosystem Inc., USA) using the appropriate profile as designed for each primer pair [11,12].
Molecular Integrity of bacteria isolates
The integrity of the amplified gene fragments was checked on a 1.5% Agarose gel ran to confirm amplification. The buffer (1XTAE buffer) was prepared and subsequently used to prepare 1.5% agarose gel. The suspension was boiled in a microwave for 5 minutes. The molten agarose was allowed to cool to 60°C and stained with 3μl of 0.5 g/ml ethidium bromide (which absorbs invisible UV light and transmits the energy as visible orange light). A comb was inserted into the slots of the casting tray and the molten agarose was poured into the tray. The gel was allowed to solidify for 20 min to form the wells. The 1XTAE buffer was poured into the gel tank to barely submerge the gel. Two microliters (2 l) of 10X blue gel loading dye (which gives color and density to the samples to make it easy to load into the wells and monitor the progress of the gel) was added to 4μl of each PCR product and loaded into the wells after the 100bp DNA ladder was loaded into well 1. The gel was electrophoresed at 120V for 45 min visualized by ultraviolet trans-illumination and photographed. The sizes of the PCR products were estimated by comparison with the mobility of a 100bp molecular weight ladder that was ran alongside experimental samples in the gel [9,11].
RESULTS (Table 1)
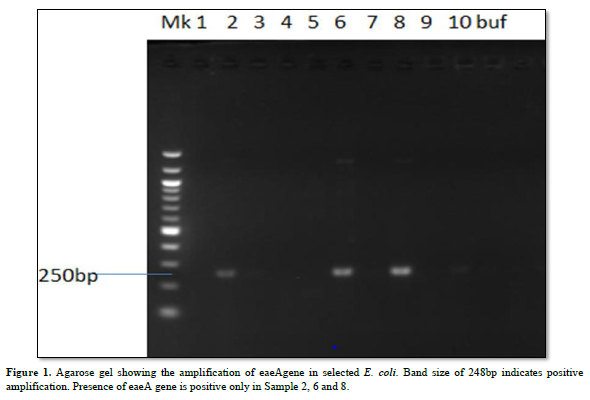
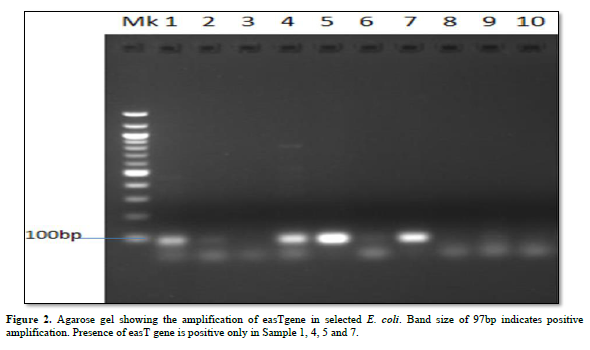
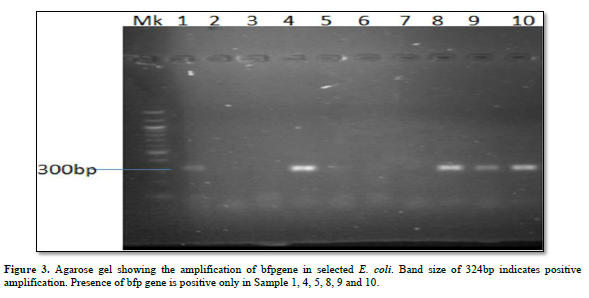
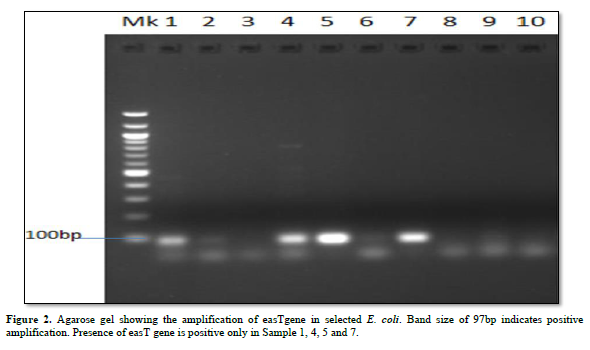
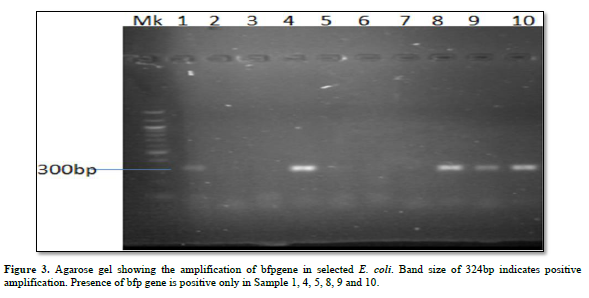
Characterization of Virulence Gene analysis from fifteen isolates under investigation were depleted in Figures 1-3, showing the Agarose gel showing the amplification of eaeAgene, bfpgene and bfpgene in selected E. coli. Band size of 248bp, 97bp and 324bp indicates positive amplification for all three virulent gene in eaeAgene (Sample 2, 6), easTgene (8, 1, 4, 5) and bfpgene (7. 1, 4, 5 and 7) respectively.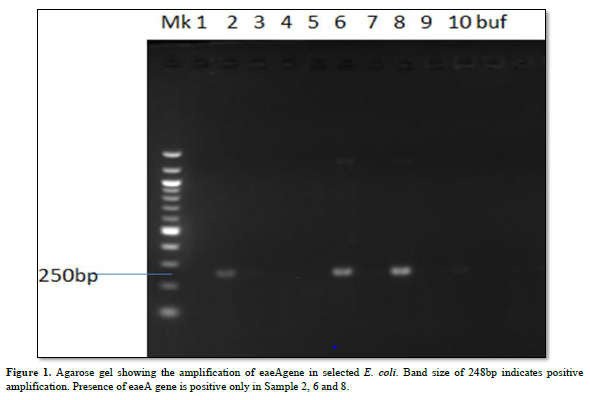
DISCUSSION
The aim and objective of this research work are to determine characterization of virulence gene in E. coli organisms isolated from slaughtered beef using 16s rRNA molecular sequencing method of analysis. Escherichia coli are an emerging and reemerging cause of food-borne infection. An estimated 10,000 to 20,000 cases of infection occur in the in Nigeria each year. Infection often leads to diarrhea, and occasionally to kidney and liver failure. Most enteric infection has been associated with eating undercooked, contaminated Slaughtered Beef under the investigation of this research work, E. coli is naturally found in the intestinal tracts of many farm animals, including healthy cattle, sheep, goats and etc., as a commensal organism ready to harm if opportunity arises [13].
The discovery of diarrheagenic E. coli in cow meat indicates a critical role as a reservoir for strains that transmit disease from animals to humans. Food produced from such animals as cows may be contaminated with DEC at the slaughterhouse, processing facility, or the consumer’s kitchen. DEC infection may also come from the consumption of raw and unpasteurized milk and vegetables contaminated with feces [14].
Diarrheagenic E. coli (DEC) (is one of the first pathogens to be observed using today’s advanced molecular diagnostic of 16S rRNA molecular gene sequencing methods. These methods are the most popular and reliable in differentiating E. coli (DEC) strains from those of non-enteric pathogenic bacteria. Moreover, the phylogeny and taxonomy of bacteria can also be studied using 16S rRNA gene sequence as differential genetic markers [6,15-18].
The gene molecular sequence of 16S rRNA is used because the 16S rRNA gene is found in almost all bacteria, the 16S rRNA gene does not change its function over time, and the 16S rRNA gene (1500 bp) is large enough for information purposes, one of the most interesting potentials of 16S rRNA gene is its ability to provide genus and species identification for isolates and also predetermined the virulent gene adherent in the gene clusters of the cell. This virulent gene plays an important role in the ability of E. coli to infect and establish infection and diseases [19-21]. The effectiveness of 16S rRNA should be stressed as a matter of urgency, Characteristics of molecular targets from these 16S rRNA methods allow the study of bacterial phylogenetics, both for bacteria detection or identification in clinical laboratories [22,23]. It is difficult to overlook the activity of the virulent gene under the scope of this research. The easTgene is expressed in the neurogenic region at the time of neuroblast segregation and in cells in the peripheral and central nervous system of E. coli [24], this is one focal point of this research work.
However, the pathogenesis of E. coli should be mentioned, for understanding the activity of these virulent genes. Escherichia coli (EPEC) strains were the first E. coli strains associated with gastroenteritis and continue to be a leading cause of diarrhea. pathogenesis of EPEC infections and other E. coli strains three-stage model [25]. We can mention one or two stages under the scope of this research, for clarity purposes.
According to a model, postulated by a researcher [26]. The first model initial adherence of the (Escherichia coli) organism to the epithelial cell, recognized in tissue cultures as localized adherence, is characterized by the formation of micro-colonies on the cell surface and is dependent on a large plasmid common to Escherichia coli strains [26]. The second model postulated is the initial contact, the bacterium transduces, a signal to the epithelial cell that results in the activation of host cell tyrosine kinase activity [27], and the third model postulated is the elevation of intracellular calcium concentrations [28]. E. coli becomes intimately attached to the membrane of the epithelial cell, with damage to host cell microvilli and accumulation of cytoskeletal proteins beneath the adherent organism [29]. The latter two steps are collectively referred to as attaching and effacing Subsequent to these events, a subset of the bacteria enters the epithelial cell of host tissues.
However, there is a need to mention the virulent activity of the characterized gene responsible for the virulent factor in the species of E. coli under the scope of this research work. eaeA gene, initially termed eae, isolated as a locus this gene is necessary for the attaching and effacing activity of E. coli. Sighting an example in EPEC [30]. This eaeAgene is found on the chromosome of E. coli strains. The novel gene is responsible display attachment activity of all E. coli strain, which gives E. coli the ability to cause rapid infection [31,32]. eaeA gene for required intimate attachment to host epithelial cells and identification of this novel gene is a function of virulence determinant of E. coli strains. We can say that eaeA gene is the second chromosomal gene necessary for the virulent capability of E. coli.
Biologists use GFP as a marker protein. Gfp refers to the gene that produces green fluorescent protein. Fluorescent proteins help whole-body imaging of tumors on internal organs. These multicolored proteins have allowed the color-coding of cancer cells to grow in vivo with the distinction of different cell types, including host from the tumor, with single-cell resolution. Photoactivatable fluorescent proteins enable the tracking of photo labeled molecules and cells in space and time and can also be used for super-resolution imaging. Gfp can be termed genetically encoded sensors which makes it possible to monitor the activity of enzymes and the concentrations of various analytes [33].
The major disadvantage of studying GFP fusion proteins is that they are generally over-expressed relative to endogenous proteins. The GFP tag can, in principle, affect protein function which makes them more predominantly virulent genes, two advantages of GFP as a reporter of gene expression are that I protein accumulation can be directly observed in living cells prior to quantitative analysis, and ii. GFP gene expression can be measured on a cell-by-cell basis [34].
BFP gene is the third virulent gene under the scope of this research work. They completely cleaved into the 201 and 123 bp fragments, and no 324 bp fragment remained in 16S rRNA sequencing profiling. We should mention the base pair which the gene function properly with positive amplification it was observe that the function between 97bp to 324bp. (easTgene band size of 97bp, eaeAgene 248bp and bfpgene 324bp respectively in Figures 1-3, 324 bp band was observed due to the editing where both the enzyme and gRNA were applied together, 201 bp and 123 bp in the same sample of the same lane where restored GFP was expressed (Figure 3) we could see the cleaved bands which restored for genetic code is not possible which we have also observed from our confocal images. when an expression vector is introduced into the cell, an endogenous transcription factor binds to the vector promoter, Therefore, the transcription of endogenous genes is totally suppressed. Necessary transcription factors are depriving, the easTgene was found in E. coli strains they are detected more frequently in strains of E. coli, and other E. coli, family mention above. This suggests that their toxin is insufficient to cause diarrhea unless another virulence factor is present as an adjuvant which is demonstrated in this research work. These results agree with those reported by Savarino [35] who found the astA gene in a significant proportion of E. coli intestinal isolates from children without diarrhea in developing countries [35]. For this gene to function properly, the novel gene must work in conjunction with other novel gene like BFP, eaeA gene and etc. if we find one, other can also present in the race to make E. coli more virulent.
CONCLUSION
In conclusion, E. coli strains have received increased attention as a cause of human diarrhea in many populations. However, the virulence factors that characterize this type of diarrheagenic E. coli are not well known but with the advent of 16S rRNA molecular sequencing, it was deduced that easTgene, eaeA gene, and bfpgene novel genes are responsible for the virulent capability of E. coli strain present in Slaughtered Beef. This has been proved beyond all reasonable double and extra caution should be taken in the Slaughtered house to prevent Slaughtered Beef contamination, and to reduce the scourge of E. coli and its other species infestation.
- Pitout JDD (2012) Extraintestinal pathogenic Escherichia coli: A combination of virulence with antibiotic resistance. Front Microbiol 3: 9.
- CDC (2014) Questions and answers coli. Available online at: https://www.cdc.gov/ecoli/general/index.html
- Donnenberg MS, Whittam TS (2001) Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J Clin Invest 107: 539-548.
- Wang L, Liu N, Gao Y, Liu J, Huang X, et al. (2021) Surveillance and Reduction Control of Escherichia coli and Diarrheagenic coli During the Pig Slaughtering Process in China. Front Vet Sci 8: 735076.
- Blanco M, Blanco JE, Mora A, Dahbi G, Alonso MP, et al. (2004) Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from healthy sheep in Spain and identification of a new intimin variant gene (eae-ξ). J Clin Microbiol 42(2): 645-651.
- Botkin DJ, Galli L, Sankarapani V, Soler M, Rivas M, et al. (2012) Development of a multiplex PCR assay for detection of Shiga toxin-producing Escherichia coli, enterohemorrhagic coli, and enteropathogenic E. coli strains. Front Cell Infect Microbiol 2(8): 1-10.
- Montiel-González MF, Vallecillo-Viejo IC, Rosenthal JJ (2016) An efficient system for selectively altering genetic information within mRNAs. Nucl Acids Res 44(21): 157-168.
- Indraswari A, Haryanto A, Suardana IW, Widiasih DA (2021) Isolation and detection of four major virulence genes in O157: H7 and non-O157 coli from beef at Yogyakarta Special Province, Indonesia. J Anim Heal Prod 9: 371-379.
- Kanamori H, Navarro RB, Yano H, Sombrero LT, Capeding MRZ, et al. (2011) Molecular characteristics of extended-spectrum β-lactamases in clinical isolates of Enterobacteriaceae from the Philippines. Acta Trop 120: 140-145.
- Osuntokun OT, Azuh VO, Adejoro BF, Akele EO (2021a) Antimicrobial Spectrum, Growth/Killing Kinetics, Conventional/Molecular Assay of Characterizing Non-Leguminous Endophytic Bacteria and Fungi from Helianthus annuus, Carica papaya and Lycoperesicum solanum. J Biomed Res Environ Sci 2(10): 1018-1034.
- Osuntokun OT (2021b) Antimicrobial Spectrum, Growth/ killing kinetics, Conventional /Molecular assay and Ultraviolet Spectrophotometer Signatures of Characterizing Shigella Flexneri and Enterococcus Faecalis and Isolated from Swine House isolates. Int J Pharm Infect Ther 4(1): 1-27.
- Chong Y, Shimoda S, Shimono N (2018) Current epidemiology, genetic evolution and clinical impact of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Genet Evol 61: 185-188.
- Wardoyo EH, Suardana IW, Yasa IWPS, Sukrama IDM (2021) Antibiotics susceptibility of Escherichia coli isolates from clinical specimens before and during COVID-19 pandemic. Iran J Microbiol 13(2): 156-160.
- WHO (2018) Key facts pp: 1-7. Available online at: https://www.who.int/news-room/fact-sheets/detail/e-col
- Fujioka M, Kasai K, Miura T, Sato T, Otomo Y (2009) Rapid diagnostic method for the detection of diarrheagenic Escherichia coli by multiplex PCR. Jpn J Infect Dis 62: 476-480.
- Amarantini C, Sembiring L, Kushadiwijaya H, Asmara W (2011) Identification and characterization of Salmonella typhi isolates from Southwest Sumba District, East Nusa Tenggara based on 16S rRNA gene sequences. Biodiversitas 12(1): 1-6.
- Fialho OB, de Souza EM, de Borba Dallagassa C, de Oliveira Pedrosa F, Klassen G, et al. (2013) Detection of diarrheagenic Escherichia coli using a two-system multiplex-PCR protocol. J Clin Lab Anal 27: 155-161.
- Suardana IW, Sumiarto B, Lukman DW (2007) Isolation and identification Escherichia coli O157:H7 on beef at Badung Regency Province of Bali. J Vet 8(1): 16-23.
- Patel J (2001) 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. Mol Diagn 6(4): 313-21.
- Pangastuti A (2006) Species definition of prokaryotes based on 16S rRNA and protein coding genes sequence. Biodiversitas 7(3): 292-296.
- Janda JM, Abbott SL (2007) 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J Clin Microbiol 45(9): 2761-2764.
- Rahmani S, Mosavp M, Rezaeej A (2006) Detection of bacteria by amplifying the 16S rRNA gene with universal primers and RFLP. Med J Islam Repub Iran 19(4): 333-338.
- Rinanda T (2011) Analysis of 16S rRNA sequences in the field of microbiology. J Kedokt Syiah Kuala 3: 172-177.
- Zhang F (2011) Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol 29(2): 149-153.
- Donnenberg MS, Kaper JB (1992) Minireview: Enteropathogenic Eschenchia coli. Infect Immun 60: 3953-3961.
- Nataro JP, Scaletsky ICA, Kaper JB, Levine MM, Trabulsi LR (1985) Plasmid-mediated factors conferring diffuse and localized adherence of enteropathogenic Escherichia coli. Infect Immun 48: 378-383.
- Rosenshine I, Donnenberg MS, Kaper JB, Finlay BB (1992) Signal exchange between enteropathogenic Escherichia coli (EPEC) and epithelial cells: EPEC induce tyrosine phosphorylation of host cell protein to initiate cytoskeletal rearrangement and bacterial uptake. EMBO J 11: 3551-3560.
- Baldwin TJ, Ward W, Aitken A, Knutton S, Williams PH (1991) Elevation of intracellular free calcium levels in HEp-2 cells infected with enteropathogenic Escherichia coli. Infect Immun 59: 1599-1604.
- Finlay BB, Rosenshine I, Donnenberg MS, Kaper JB (1992) Cytoskeletal composition of attaching and effacing lesions associated with enteropathogenic Escherichia coli adherence to HeLa cells. Infect Immun 60: 2541-2543.
- Jerse AE, Yu J, Tall BD, Kaper JB (1990) A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci USA 87: 7839-7843.
- Kain KC, Barteluk RL, Kelly MT, Xin H, Hua GD, et al. (1991) Etiology of childhood diarrhea in Beijing, China. J Clin Microbiol 29: 90-95.
- Albert MJ, Faruque SM, Ansaruzzaman M, Islam MM, Haider K, et al. (1992) Sharing of virulence-associated properties at the phenotypic and genetic levels between enteropathogenic Escherichia coli and Hafnia alvei. J Med Microbiol 37: 310-314.
- Thomas G, Shannon JS, Sai-lan S, Jia L (2016) Genome editing technologies: Principles and applications. Cold Spring Harbor. Perspect Biol 8: a023754.
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533(7603): 420-424.
- Savarino SJ, McVeigh A, Watson J (1996) Enteroaggregative Escherichia coli heat-stable enterotoxin is not restricted to enteroaggregative coli. J Infect Dis 173: 1019-1022.